Guidelines for authors
Step-by-step guide to format and submit papers to Computo.
1 Introduction
This document provides a comprehensive documentation to accompany the authors during the writing of their contribution to Computo. It covers the requirements for submission, how to use the provided templates, how to set up dependencies, and how to ensure reproducibility through continuous integration and finally.
- Official templates (R, Python, Julia):
- Create a new GitHub repository from the appropriate template
- This contains document, environment and CI configuration files
- Local Setup:
- Clone the repository.
- Install Quarto and the Computo Quarto extension.
- Ensure you can render the manuscript locally.
- Writing Your Contribution:
- Main content goes in
template-computo-LANG.qmd. - Metadata (title, authors, abstract, etc.) is set in
_quarto.yml. - Edit using any text editor or IDE (VS Code/RStudio/Positron recommended).
- Main content goes in
- Dependencies:
- Freeze package versions for reproducibility.
- Use
renv(R),conda/requirements.txt(Python), orProject.toml(Julia). - System dependencies can be installed via
setup-env-ci.shandsetup-render-ci.sh.
- Continuous Integration (CI):
- Uses GitHub Actions via a provided
build.ymlworkflow. - Automatically builds and publishes HTML/PDF outputs to GitHub Pages.
- Uses GitHub Actions via a provided
- Publishing:
- Push changes to GitHub; CI will build and deploy the site.
- Ensure that both PDF and HTML versions render correctly.
- Submit the PDF and repository link to OpenReview.
- Ensure that the HTML is publicly accessible for reviewers on the GitHub Pages site.
2 Step-by-step guide
Submissions to Computo require both scientific content (such as equations, code, figures, and data) and clear evidence that this content is reproducible. This is achieved through: i) a notebook system, ii) a virtual environment with frozen dependencies, and iii) continuous integration. If needed, large data files can be stored externally on platforms like Zenodo or OSF.
A Computo submission is therefore a Git(Hub) repository containing:
- the source files for the notebook (a Quarto
.qmdfile, a BibTeX.bibfile, and any static files such as figures or small.csvdata tables); - configuration files to set up dependencies in a virtual environment;
- configuration files to enable continuous integration and render the final documents.
2.1 Template repositories
The Computo organization provides template repositories for R, Python, and Julia contributors.
These templates provide a starting point for authors to format their contributions according to Computo’s guidelines. They include the necessary files and configurations to set up a reproducible environment and continuous integration
2.2 Setup a git repository
Git is a versioning tool and GitHub is an online platform owned by Microsoft which provides the service of hosting repositories with all the capabilities of versioning offered by Git.
Computo requires authors to submit their contribution as a GitHub repository. Hence, authors are expected to have basic knowledge of Git. We refer authors to the following online book which is particularly well written: https://happygitwithr.com.
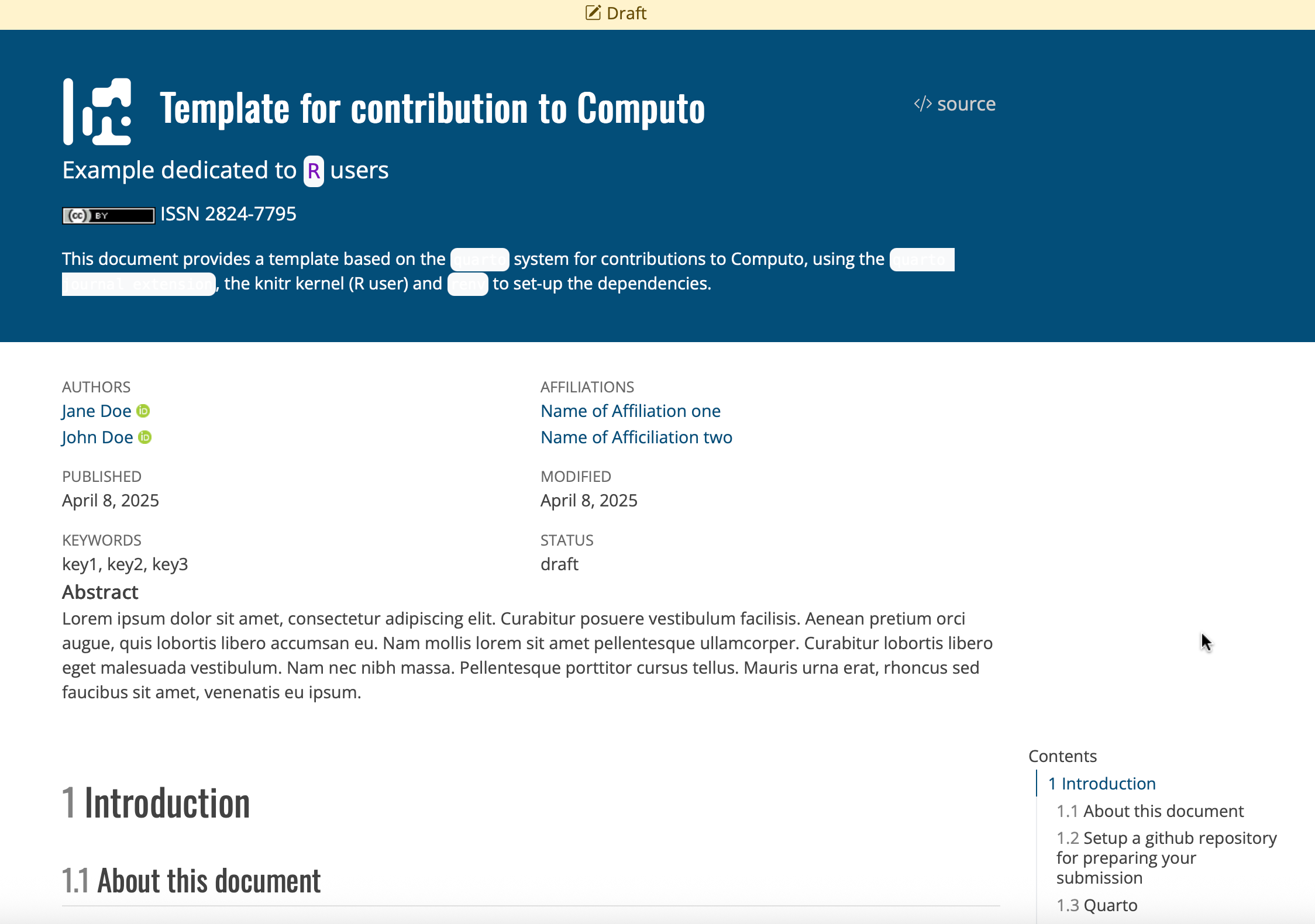
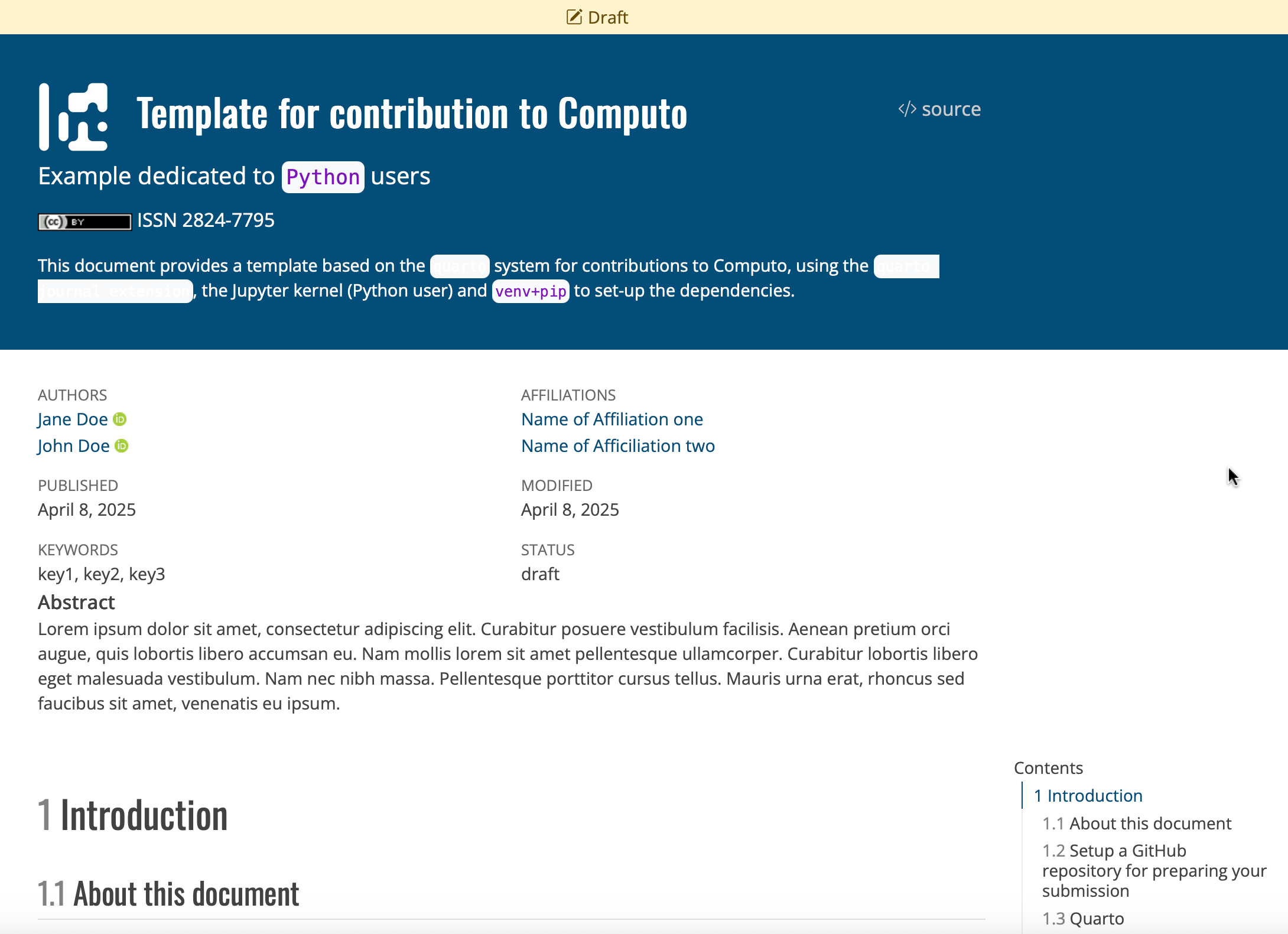
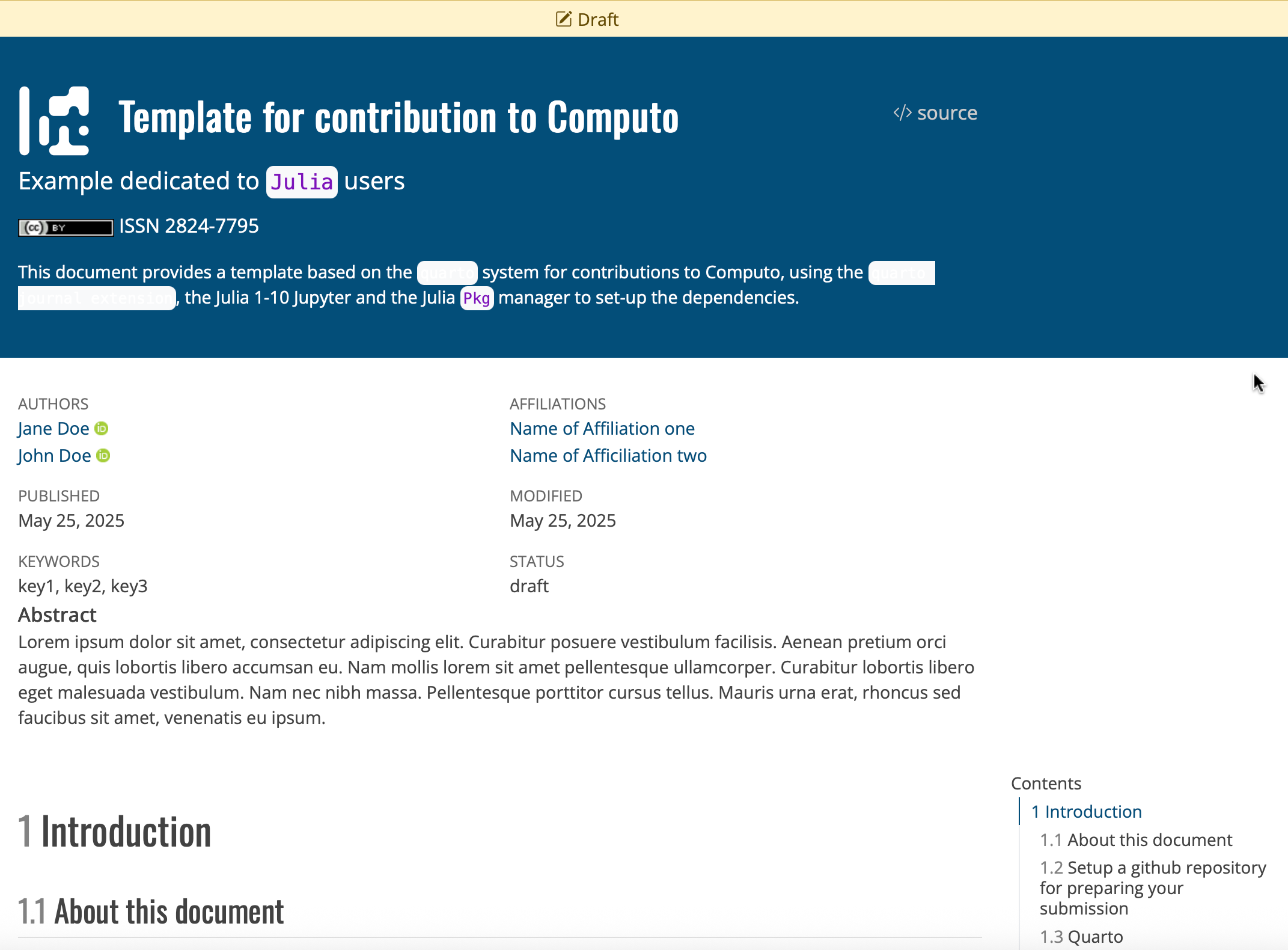
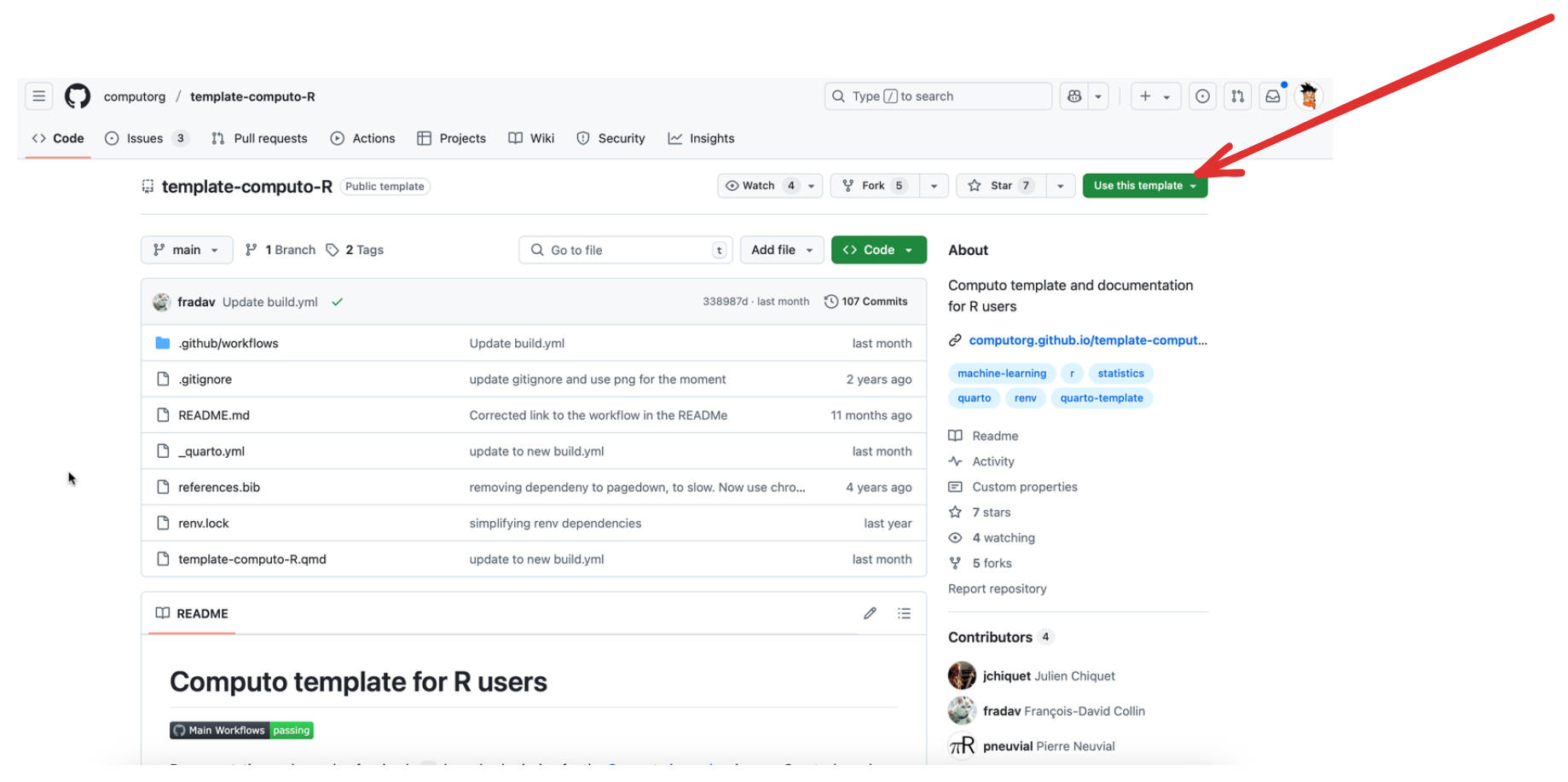
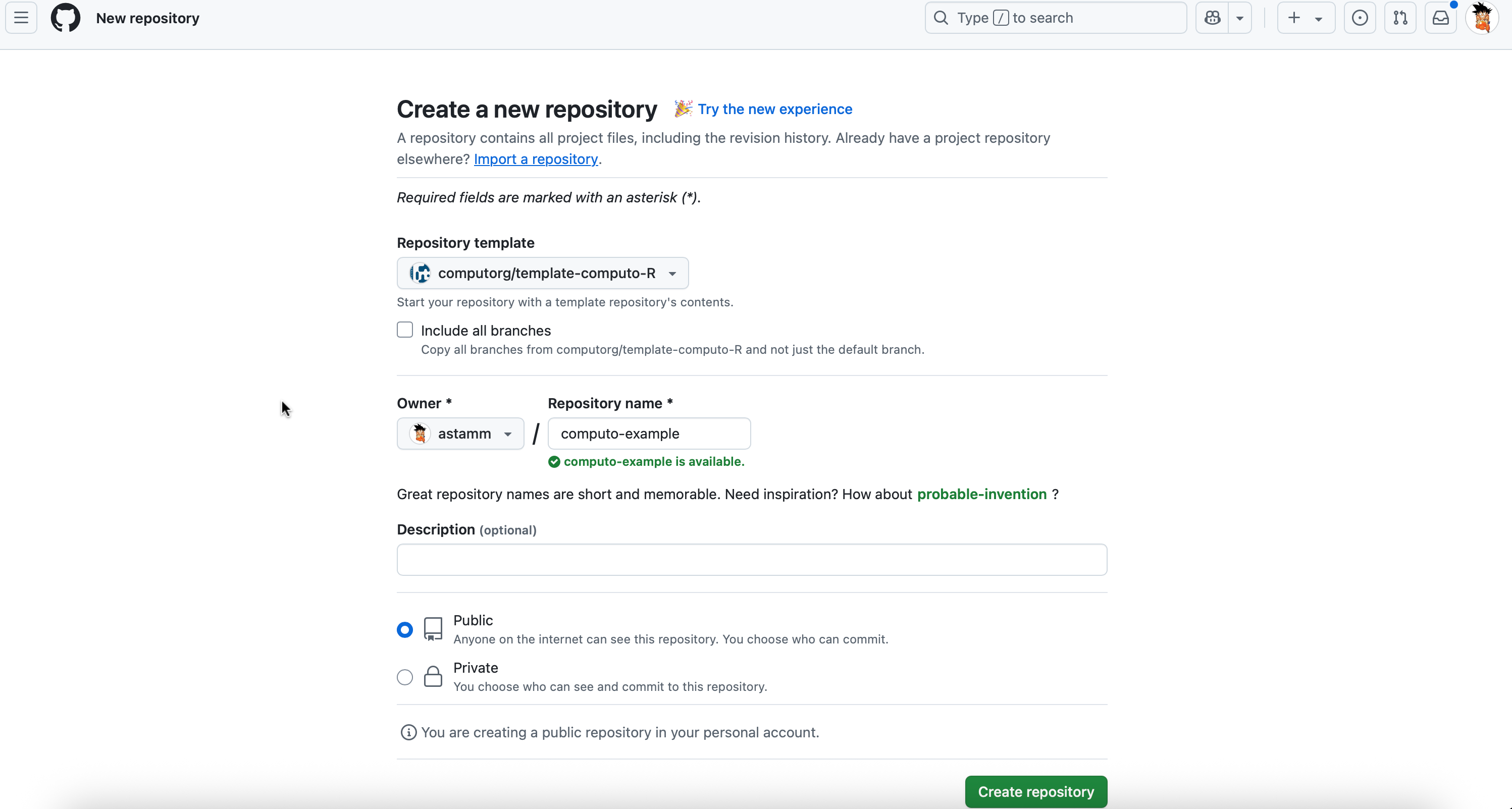
Create a new GitHub repository by navigating to the R, Python, or Julia template repository and clicking the “Use this template” button at the top of the page, as shown in Figure 1.
When you fork a GitHub repository – whether it is a classic repository or a template – GitHub will ask if you want to include all branches (there is a box labeled Include all branches). You only need to fork the main branch, so you do not need to check that box.
The paper will be automatically published online using GitHub pages. This is possible only with public repositories (which is the default behavior when you initialize your repository from this template, see Figure 2). If your prefer, it is fine to keep your repository private during the writing phase. However, we ask you to set it public at the submission stage; so that the paper can be rendered and published online for the reviewers.
You can use Gitlab for submitting to Computo. We will be giving more detailed support for this in the future.
2.3 Local setup: clone, quarto, and computo extension
First, clone the repository to your local computer. In a terminal, run:
# Clone the repository
git clone http://github.com/<user-name>/<repository-name>.gitYou need to have Quarto and the Computo extension installed.
- Install Quarto: follow instructions at https://quarto.org/docs/get-started/.
- Once Quarto is installed, open a terminal in your cloned repo folder and run:
# Install computo quarto extension
quarto add computorg/computo-quarto-extensionIf you are collaborating with others when writing your contributions, each collaborator needs to perform these same three steps (clone the repository, install Quarto and install the Computo Quarto extension).
Quarto relies on a specific engine to compile your document. By default, that engine detects code blocks, executes them and paste the result below the code block in the rendered document.
It is possible to use multiple languages within the same document (e.g. R and Python or Python and Julia, etc.) provided that you use the knitr engine. Indeed, knitr is an R package that is able to parse code blocks from many languages. This can be achieved by specifying engine: knitr in your document metadata. The default engine binding that Quarto uses is detailed here: https://quarto.org/docs/computations/execution-options.html#engine-binding.
Before proceeding to the next step, make sure that you are able to build your manuscript as a standard notebook on your system by running the following command in a terminal:
# will render both to html and PDF
quarto render2.4 Write your contribution
There are mainly two files you are expected to modify in your repository for writing your contribution: template-computo-LANG.qmd (where LANG is to be replaced with either R, Python or Julia) and _quarto.yml. While the first can be renamed (e.g, submission-YYYYMM-firstAuthor-keyword.qmd), the second must remain _quarto.yml as it contains metadata about your contribution shared between project’s files.
2.4.1 Metadata of your contribution
All yaml metadata should preferably be placed in the _quarto.yml file. To customise title, authors, date, abstract and so on, you should modify _quarto.yml which by default looks like this:
project:
type: default
render:
- template-computo-LANG.qmd
- README.qmd
title: "Template for contribution to Computo"
subtitle: "Example dedicated to `LANG` users"
author:
- name: Jane Doe
corresponding: true
email: janedoe@nowhere.moon
url: https://janedoe.someplace.themoon.org
orcid: 0000-0000-0000-0000
affiliations:
- name: Name of Affiliation one
department: Statistics
url: https://someplace.themoon.org
- name: John Doe
email: johndoe@nowhere.moon
url: https://johndoe.someplace.themoon.org
orcid: 0000-0000-0000-0000
affiliations:
- name: Name of Afficiliation two
department: Computer Science
url: https://someplace.themoon.org
date: last-modified
date-modified: last-modified
description: |
This document provides a template based on the [`quarto`](https://quarto.org/) system for contributions to Computo, using the [`quarto journal extension`](https://github.com/computorg/computo-quarto-extension), the knitr kernel (R user) and [`renv`](https://rstudio.github.io/renv/articles/renv.html) to set-up the dependencies.
abstract: >+
Lorem ipsum dolor sit amet, consectetur adipiscing elit. Curabitur posuere vestibulum facilisis. Aenean pretium orci augue, quis lobortis libero accumsan eu. Nam mollis lorem sit amet pellentesque ullamcorper. Curabitur lobortis libero eget malesuada vestibulum. Nam nec nibh massa. Pellentesque porttitor cursus tellus. Mauris urna erat, rhoncus sed faucibus sit amet, venenatis eu ipsum.
keywords: [key1, key2, key3]
bibliography: references.bib
github-user: computorg
repo: "template-computo-r"
draft: true # set to false once the build is running
published: false # will be set to true once accepted
license: CC-BY-4.0
format:
computo-html: default
computo-pdf: defaultYou can customize most of the entries in that file except project:, published:, draft:, license: and format:. If your contribution is accepted for publication, the associate editors will modify parts of this file accordingly.
2.4.2 Main content of your contribution
The main content of your contribution must be written in the template-computo-LANG.qmd file (see formatting suggestion therein). To compile your contribution, run quarto render anytime needed.
We also provide a README.qmd file as part of the project. This file uses shortcodes to extract metadata from _quarto.yml and generate a README.md that will be displayed on the main page of your GitHub repository. The README.md file is rendered automatically when you run quarto render at the root of your repository. You may modify the README.qmd file if you wish, at your own risk. If your submission is accepted, it will be standardized by the Computo team.
2.5 Setup dependencies
The next step is to inform Computo of the other packages, tools and environment that your paper might depend upon. It is important to freeze their versions to ensure reproducibility. This step is inherently handled differently whether you are an R, Python or Julia user.
2.6 Ensure reproducibility
Now that you have written your contribution in the correct template and set up its dependencies, you need to set up continuous integration via GitHub Actions in order to ensure reproducibility of your work and its publication as a webpage.
2.6.1 Use Computo’s built-in GitHub Action workflow
Computo has a built-in workflow for just that purpose which comes in the form of a YAML file describing a GitHub Action. It should have been automatically added to your repository when you cloned one of our templates. It is located under .github/workflows/build.yml.
build.yml
Authors should under no circumstances modify this file. If you encounter a problem (such as the CI not completing successfully), please get in touch with us at contact@computo-journal.org.
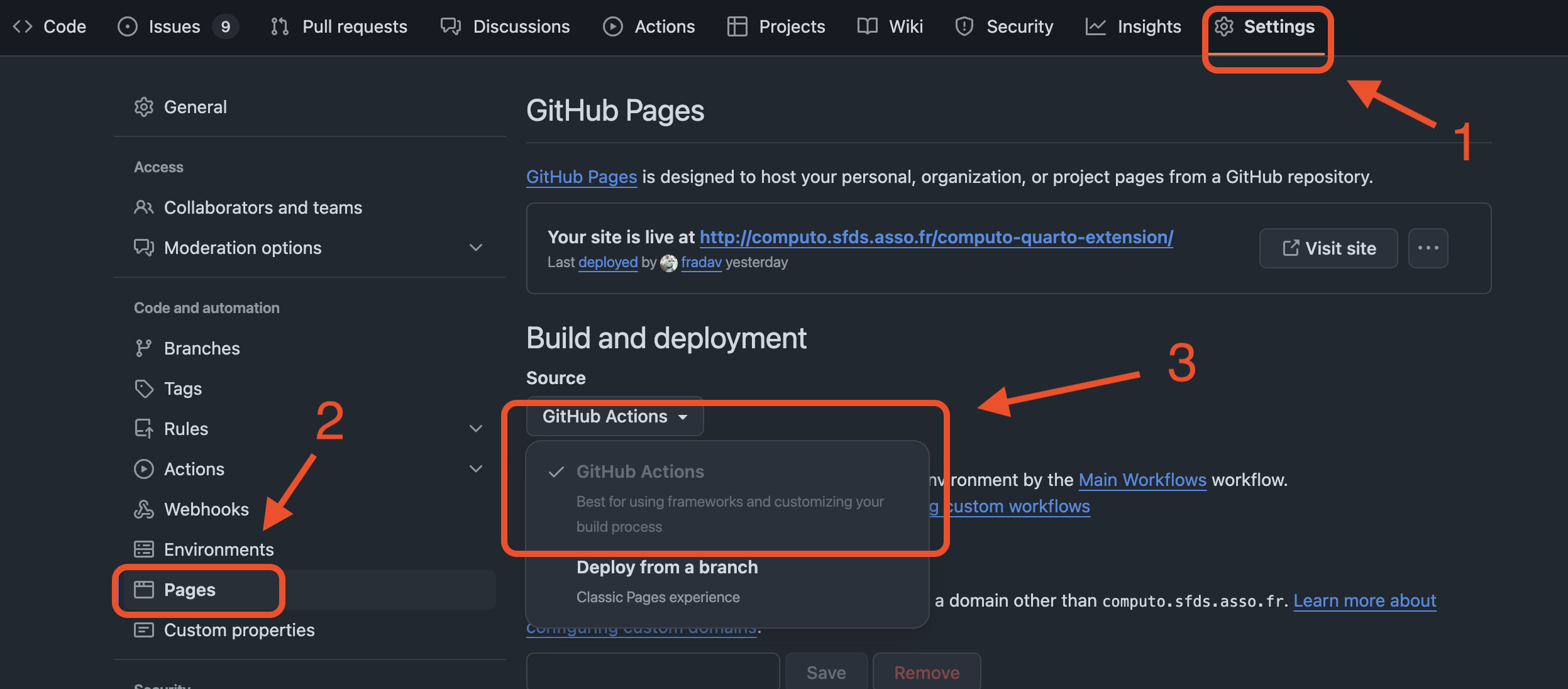
Next, as illustrated in Figure 3, under Settings > Pages on the webpage of your repository, select GitHub Actions as the source for building and deploying your website.
Authors can find more details about Computo’s workflow in the corresponding FAQ entry.
You can safely delete the gh-pages branch if you have one, as we don’t need anymore to push the HTML files to the site.
2.6.2 Handle external system dependencies
While R, Python, or Julia package dependencies should be managed using the appropriate environment tools, your project may also require external system dependencies (such as libraries or command-line tools). These must be installed on the GitHub runner during environment setup or rendering. To handle this, add and customize two scripts at the root of your repository:
setup-env-ci.shis meant to install system dependencies required to set up the environment in the GitHub runner. Here is an example of such a file taken from a published article:
# The CI runs on Linux Ubuntu, here goes system dependencies
# needed at environment setup step
sudo apt-get install -y libcurl4-openssl-devsetup-render-ci.shis meant to install system dependencies required for rendering the paper, like plotting software or running a script to generate data. Here is an example of such a file taken from a published article:
# The CI runs on Linux Ubuntu, here goes system dependencies
# needed at rendering step
sudo apt-get install -y libblas-dev liblapack-devRemember that the GitHub runner uses Linux Ubuntu and therefore software installation is achieved through apt-get. Furthermore, you must precede calls to apt-get with the word sudo to indicate that you wish to run this command as an administrator.
2.7 Push to GitHub
Once you have finished part of your writing and confirmed that the document builds successfully locally, you can push your changes to GitHub as you would with any Git project. If everything is set up correctly, the continuous integration workflow will automatically build and publish the HTML and PDF versions online.
Make sure to check and update the .gitignore file in your repository. In particular, if you changed the name of some files in the repository (like the main .qmd file), make propagate this change to the .gitignore file in order to avoid pushing binary files to the repository (such as the PDF file).
2.8 Submit your manuscript
Once you are satisfied with your manuscript, you can submit it for review. To do so, upload the generated PDF to the OpenReview platform and indicate the corresponding Git repository. This will initiate both the scientific and reproducibility review processes.
Please also ensure that the HTML version of your manuscript is publicly accessible for reviewers on the GitHub Pages site associated with your repository. To do so, complete the description of your repository (below the “about” section) by ticking the box labeled “Use your Github page Website” (see image below).
For more details about the review process, see the guidelines for reviewers.
2.9 Accepted manuscript
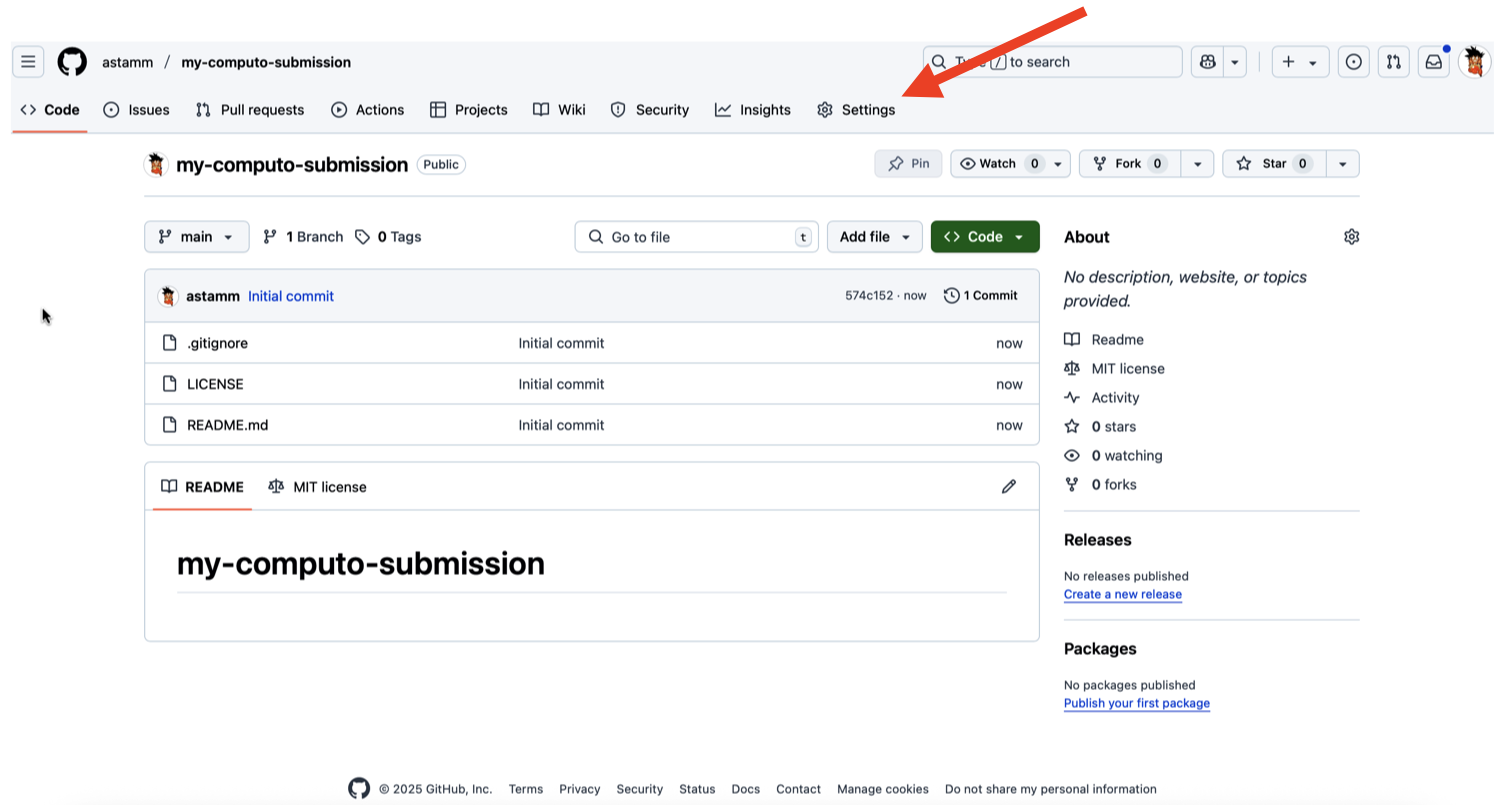
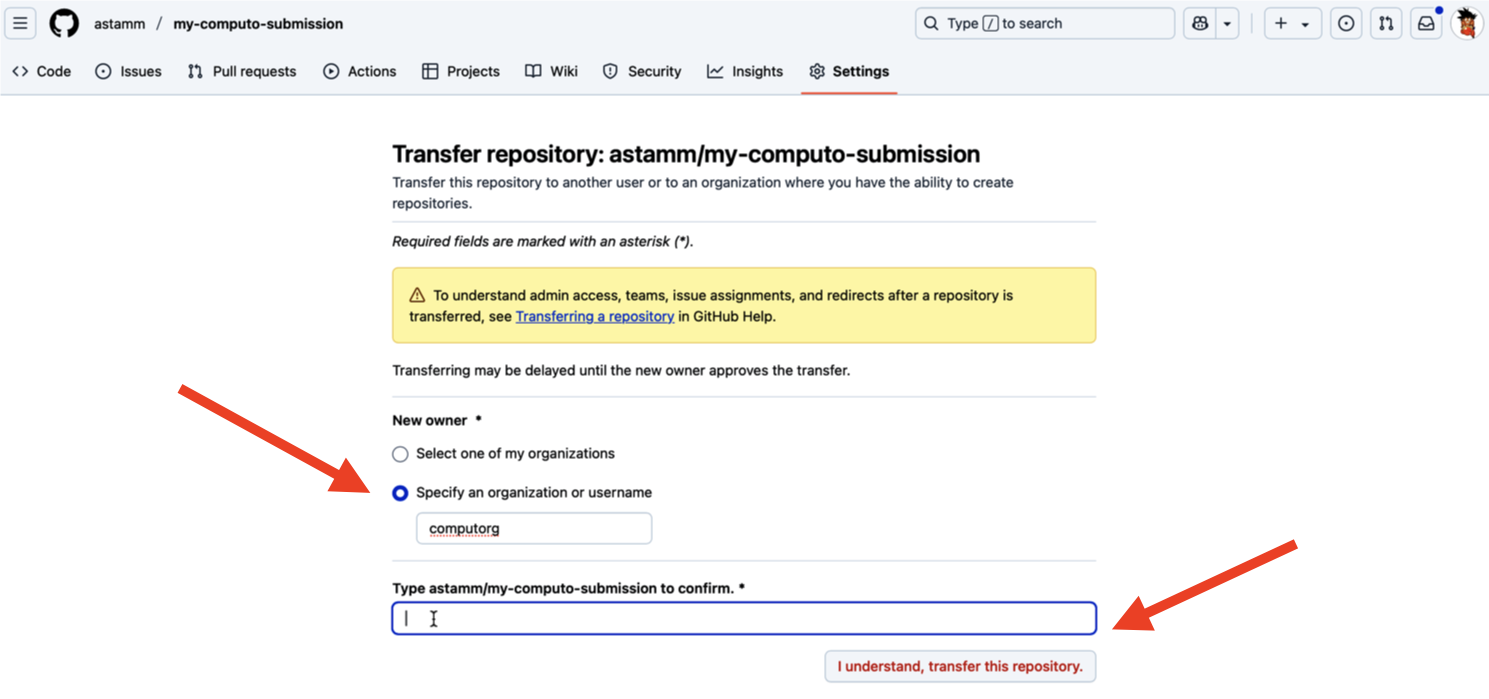
If your submission is accepted for publication, you will be asked to transfer the ownership of the repository that hosts your paper to the Computo GitHub organization. To do that, you must go on the main page of your repository in a web browser and click on the Settings tab as indicated in Figure 5.
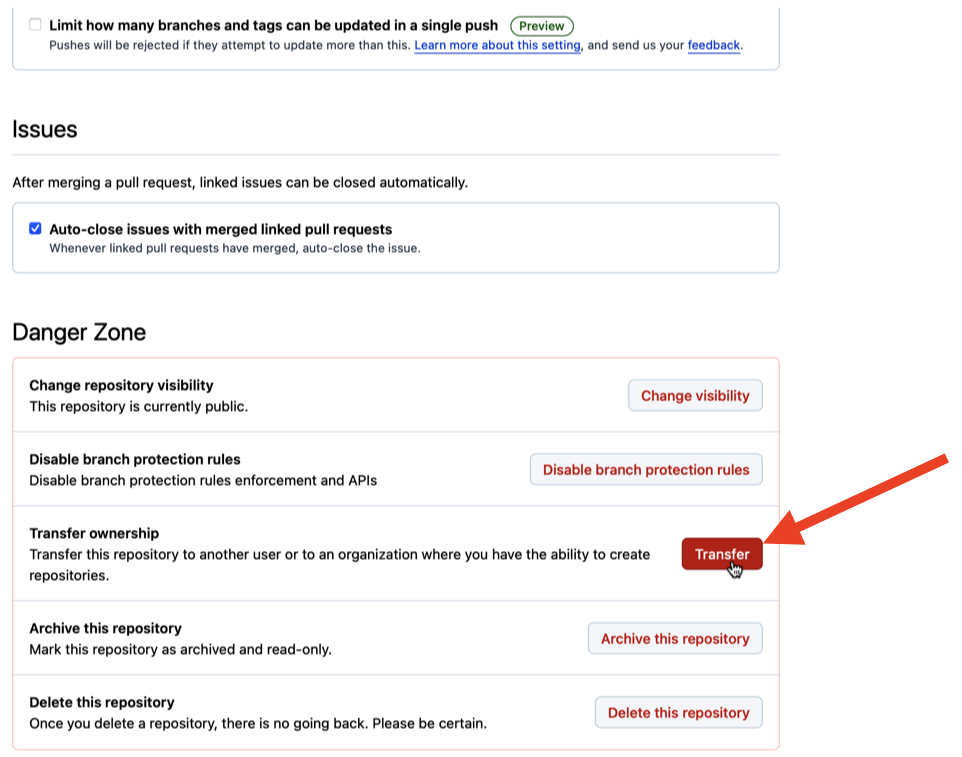
Once inside the settings of the repository, scroll all the way down to the bottom of the page until you see Danger Zone in red color and click on Transfer as indicated in Figure 6.
A new page opens where you need to select Specify an organization or username and write computorg (see Figure 7). Then you type what is asked in the blank box and click on I understand, transfer this repository and you’re all set!
3 Advanced examples
3.1 Mock contributions
In addition to the template repositories that can serve as example papers (see Section 2.1), we provide more in-depth illustrations of the expected content and available features through mock contributions consisting of a complete reworking of the famous t-SNE article.
2008
Visualizing Data using t-SNE (mock contributon)
Laurens van der Maaten and Geoffrey Hinton
Visualizing Data using t-SNE (mock contributon)
Laurens van der Maaten and Geoffrey Hinton
3.2 Previously published papers
For other advanced examples, do not hesitate to explore the source code of any paper already published in Computo to see how the template is used in practice. In particular, reproducing an existing paper published in Computo can be easily done as detailed in the FAQ.
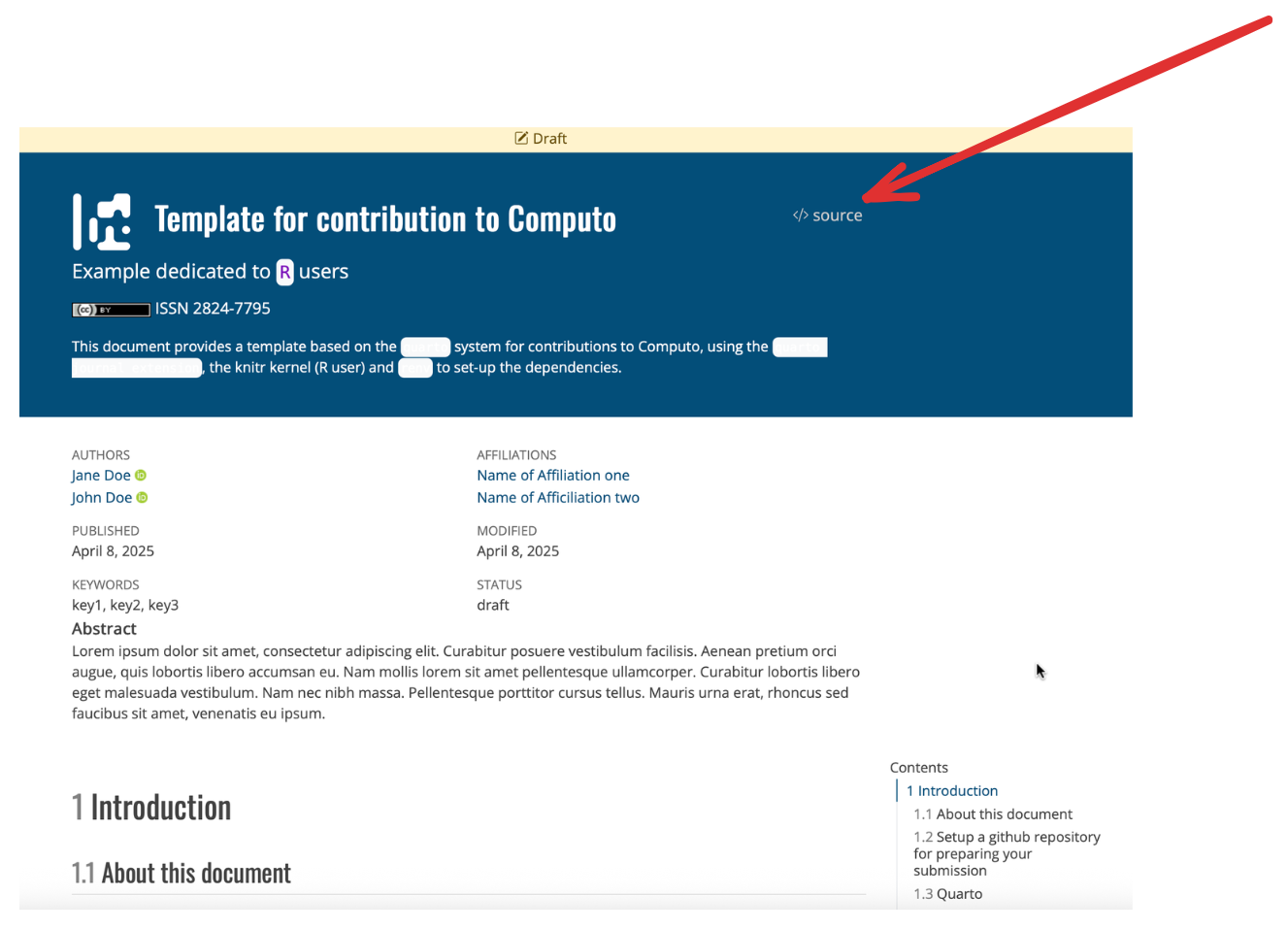
You can also access the source code of the example papers mentioned above by clicking on the </> Source button in the upper-right corner of the page as illustrated in Figure 8.
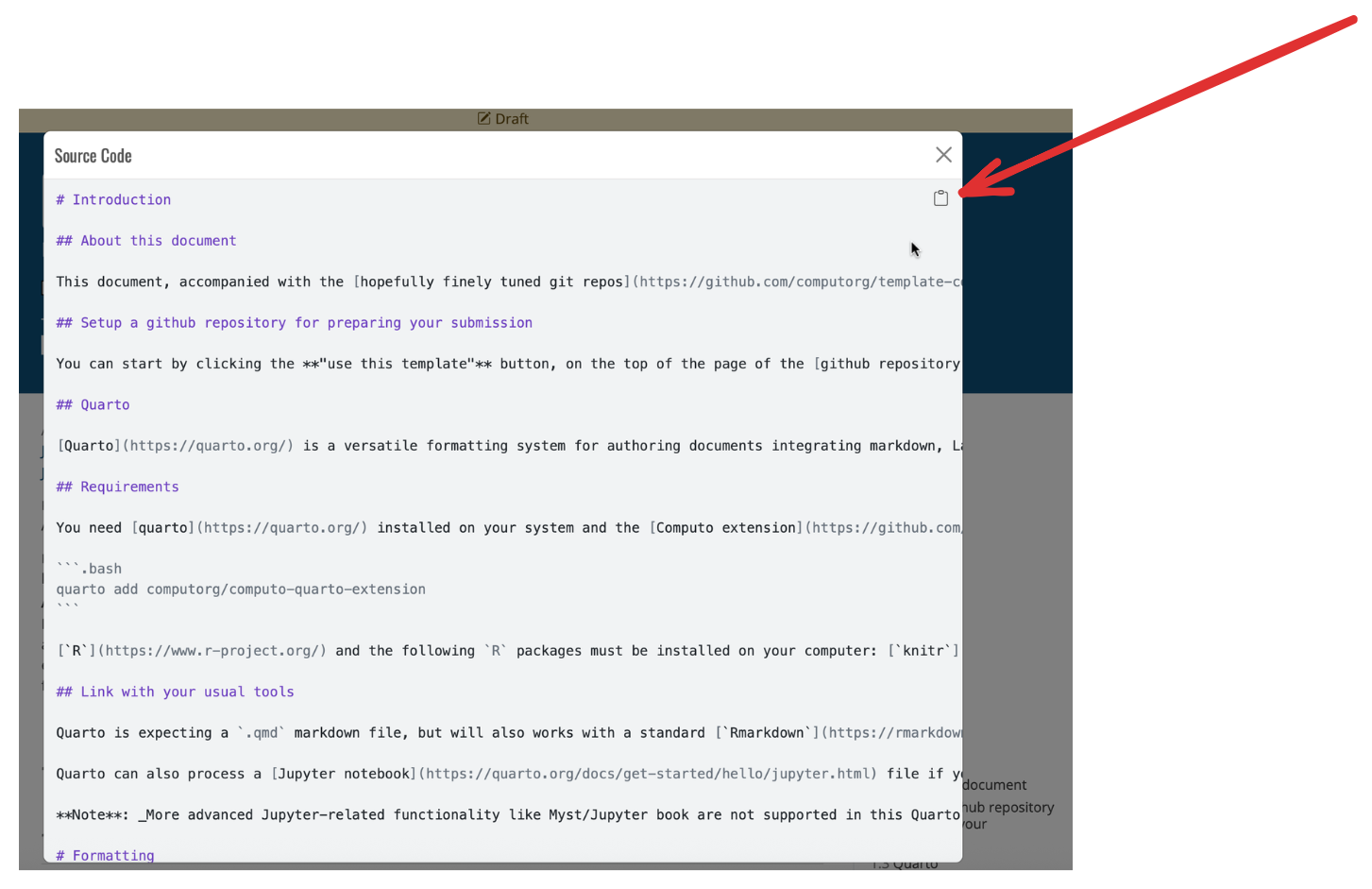
The source code of the page then displays as shown in Figure 9 and the user can click on the icon in the upper-right corner to copy the source code for practicing locally.
4 Computo’s code of ethics
- Originality
-
- Authors guarantee that their proposed article is original, and that it infringes no moral intellectual property right of any other person or entity.
- Authors guarantee that their proposed article has not been published previously, and that they have not submitted the proposed article simultaneously to any other journal.
- Conflicts of interest
-
- Authors shall disclose any potential conflict of interest, whether it is professional, financial or other, to the journal’s Editor, if this conflict could be interpreted as having influenced their work.
- Authors shall declare all sources of funding for the research presented in the article.
- Impartiality
-
All articles are examined impartially, and their merits are assessed regardless of the sex, religion, sexual orientation, nationality, ethnic origin, length of service or institutional affiliation of the author(s).
- Funding
-
All funding received by the author(s) shall be clearly stated in the article(s).
- Defamatory statements
-
Authors guarantee that their proposed article contains no matter of a defamatory, hateful, fraudulent or knowingly inexact character.
- References
-
Authors guarantee that all the publications used in their work have been cited appropriately.
- Copyright/author’s right/license compliance
-
Authors guarantee that they comply with the usage license of any third party contents/works (code, software, data, figures/images, documents, etc.) that were used to produce their work.